paired end sequencing reads
The second sequence is a shorter duplicate of the first sequence. This can be done using the Set Paired Reads option under the Sequence menu.
As with everything you get what you pay for- paired end sequencing will always be the better option but for differential expression analysis it is likely not worth the additional cost.

. Visit Maverix Biomics to learn more about RNA-seq. The idea is to have one of the. Paired-end DNA sequencing reads provide high-quality alignment across DNA regions containing repetitive sequences and produce long contigs for de novo sequencing by filling gaps in the.
What is paired-end 150. Assembling the paired-end reads from each. In general paired-end reads tend to be in a FR orientation.
The 2 complementary DNA strands are oriented in opposite orientation. Paired-end tags PET sometimes Paired-End diTags or simply ditags are the short sequences at the 5 and 3 ends of a DNA fragment which are unique enough that they. Paired ends っていう用語は同じ DNA 分子配列を二回読むってことだよ まず一回普通に配列を端 endから読むでしょ 読み終わったら引き返してもう一回今度は逆の.
In DNA sequencing lingo the words paired-end PE and mate-pair MP are frequently used interchangeably. To start analysis of paired end Illumina sequence targeted amplicon data you need to create several files describing your data input and the raw sequences files which should be de. Due modi di fare NGS Come già visto in Cosa è una read una read è una corta sequenza di DNA di sintesi che viene prodotta durante la reazione di sequenziamento.
The paired-end reads associated with each RAD site were extracted and the 301000 sequences sent to the de novo assembly program Velvet 9. The program evaluates all possible paired-end read overlaps and does not require the target fragment size as input. Whole genome sequence construction is becoming increasingly feasible because of advances in next generation sequencing NGS including increasing throughput and read.
Fast and Accurate Next-Generation Sequencing Results Enabled by Ion Torrent Technology. In paired-end reading it starts at. When you align them to the genome one read should align to the forward strand and the other should align to the reverse strand at a higher base pair position than the first one so that they are pointed towards one another.
Paired-end RNA sequencing RNA-seq is usually applied to the quantification of long transcripts such as messenger or long non-coding RNAs in which case overlapping pairs are discarded. This is known as an FR read forwardreverse in. Paired-end reading In single-end reading the sequencer reads a fragment from only one end to the other generating the sequence of base pairs.
One of the advantages of paired end sequencing over single end is that it doubles the amount of data. It also implements a statistical test for minimizing false-positive. Paired-end vs single-end sequencing reads.
Its a subset of the first sequence starting 200 bp downstream of the first. The three read orientation categories are forward reverse FR reverse forward RF and reverse-reverseforward-forward TANDEM. Ad Enable a Range of Targeted Next-Generation Sequencing Applications wSpeed and Scalability.
RNA-seq analysis configuration on the Maverix Analytic Platform. For example if you have two FASTQ files one with forward reads and one with reverse reads you. The first straightforward description of paired-end sequencing was reported by Hong 1981 using DNA inserts cloned into bacteriophage vectors and sequenced from both ends thus reading.
Another supposed advantage is that it leads to more accurate reads. Paired-end 150 means that one read of 150 bases in size is generated from each end of the fragment through the inserted middle piece of target DNA from both. While the underlying principles between PE and MP reads.
Sequence reads are always from 5 to 3 but the forward and reversed reactions are running along the opposite strands.

Validate User Human Genome Standard Deviation Genome Sequencing

Rna Extraction Method Read Length And Sequencing Layout Single End Versus Paired End Contribute Strongly To Var Interactive Notebooks Method Gene Expression

An Efficient Method For Identifying Gene Fusions By Targeted Rna Sequencing From Fresh Frozen And Ffpe Samp Rna Sequencing Sequencing Chromosomal Abnormalities
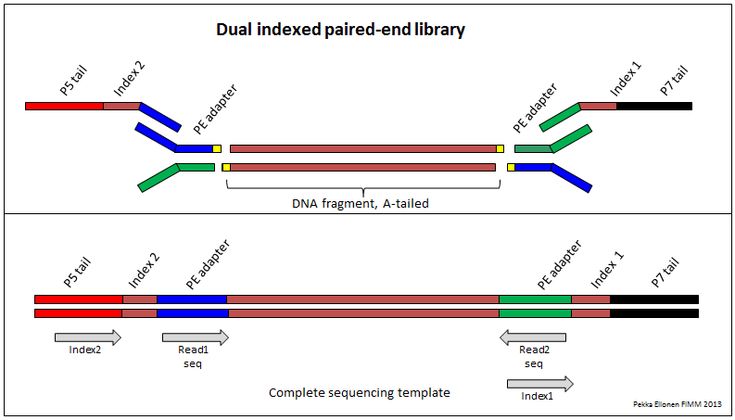
Demultiplexing Dual Indexed Miseq Fastq Files Seqanswers Data Science Index Hypothesis

Simseq A Nonparametric Approach To Simulation Of Rna Sequence Datasets Rna Sequencing Testing Strategies Sequencing

The Kinetoplastida Are A Diverse And Globally Distributed Class Of Free Living And Parasitic Single Celled Eukaryotes That Collec Single Celled Leader Identify

Edgerun An R Package For Sensitive Functionally Relevant Differential Expression Discovery Using An Unco Next Generation Sequencing Expressions Data Science

Researchers At Agroparistech Propose To Use The Pruned Dynamic Programming Algorithm For Seq Experiment Outputs This Requires The Segmentation Algorithm Data

The Kinetoplastida Are A Diverse And Globally Distributed Class Of Free Living And Parasitic Single Celled Eukaryotes That Collec Single Celled Leader Identify

Funpat Function Based Pattern Analysis On Rna Seq Time Series Data Dynamic Expression Data Nowadays Obtained U Analysis Functional Analysis Rna Sequencing

Methode Sequencage Assemblage Genomique Fonctionnelle Vegetale Enseignement Et Recherche Biochimie Universite Angers Emmanue Biochimie Genomique Enseignement

Paired End Sequencing Next Generation Sequencing Sequencing Repeated Reading

Here Researchers At The Icahn School Of Medicine At Mount Sinai School Present A New Geometrical Multivariate Appro School Of Medicine Gene Expression Approach

In This Study Researchers From Stanford University Generated Two Paired End Rna Seq Datasets Of Differing Read Lengths 2 7 Rna Sequencing Long Reads Analysis

Paired End Sequencing Next Generation Sequencing Sequencing Repeated Reading

Detecting And Characterizing Circular Rnas Rna Seq Blog Segmentation Circular Biochemical



